In my previous post, I wrote about the Phase Locking Value statistic that can be used to quantify neural synchrony between two electrodes and how it can be a proxy for connectivity. Once you have quantified changes in connectivity using such measures (others include cross spectrum, coherence), it is good to visualize your results. The connected topoplot function is just the right thing for the job.

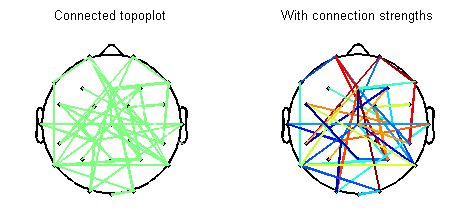
Below is a visual summary of what the script can do. In this figure, I have randomly connected ~10% of all possible channel pairs. It is possible to either specify or skip connection strengths. Detailed help and demo are available with the script.
Now, an illustration from real data. You can use the PLV statistic to isolate stimulus-induced transient networks in your EEG data. Below is typically how your results would look like – a network within a time window along with an associated connectivity pattern for the network.

In the above example, the subjects viewed a stimulus at time 0 and there are two prominent transient networks that could be isolated – one within the first 300 ms of viewing the stimulus and second between 300 and 600 ms of viewing the stimulus. Plots below the connected topoplots are an average PLV time course of the whole network that is visualized. Therefore, in the first 300 ms, the isolated network de-synchronizes and between 300 ms and 600 ms, (a different) isolated network synchronizes.
Good luck with your analyses!
Hi,
thanks for sharing your script. i run pn_tc_demo file and got the following error message:
??? Undefined function or method ‘mtimes’ for input arguments of type ‘struct’.
Error in ==> topoplot_connect at 60
Th = pi/180*Th; % convert degrees to radians
Error in ==> pn_tc_demo at 70
topoplot_connect(ds, sample_chanlocs);
??? Error: File: topoplot_connect.m Line: 49 Column: 7
Expression or statement is incorrect–possibly unbalanced (, {, or [.
Error in ==> pn_tc_demo at 70
topoplot_connect(ds, sample_chanlocs);
I think that the error is coming from this line 49 and 51 in topoplot_connect.m
[~, ~, Th Rd indices] = readlocs( loc_file,’filetype’,’loc’);
however, let us know to to correct it
thanks
Hi Mario, thank you for your question. Please make sure you’ve installed EEGLAB. connected_topoplot.m is merely an extension of a script from EEGLAB. Also, please note that I developed this script when I was using EEGLAB 6.01b, but it should work with the newer releases too. Let me know how it goes.
Cheers,
Praneeth
Thanks Praneeth for prompt reply!
I have EEGLAB v6.03b. however, by replacing the following command
[~, ~, Th Rd indices] = readlocs( );
with
[aa,bb, Th, Rd, indices] = readlocs( );
cods will run without any errors!
Oh, I see, Thank you for letting me know. I think it depends on the version of Matlab you are using. Which version are you using?
MATLAB Version 7.7.0.471 (R2008b)
Dear Praneeth, thank you very much for your efforts, it is veryhelpful.
My question is, How Can I use the plv values (800 * 32*32), -time series with length 800 and 32 channels- to have a connectivity map in a specific time period? Sould I average values in this period or there is another way?
Thank you in advance.
All bests,
Mahmoud.
Dear Mahmoud, I’m glad you like my work.
Yes, I would average the PLV within a time window. Selecting the time window is the tricky part. You can choose to simply downsample the PLV time course, or use a moving window average, or if you have a specific hypothesis based on knowledge from other sources, then you can average the PLV time course within that time window to plot the connectivity map.
I have often found it useful to make an animation of the connectivity using the connected topoplot function that I’ve posted in this blog – sometimes it is useful to run this map on a thresholded matrix of PLV time series. The threshold can be determined by computing the PLV time series from random segments of EEG signals and then setting the threshold (individually for each channel pair) to be 3 times the standard deviation of the values from PLV time series computed from random segments of EEG signals – make sure you take the same number of random segments as the number of trials you had when you computed your PLV time course for an event.
Hope this helps.
Praneeth
Dear Praneth,
I am interested to plot (among all channels) task related coherence using topoplot_connect.m file The values for task related coherence are both negative and positive. While plotting with the topoplot_connect.m, negative values are plotted as an absolute value. In the line 63 of the m file (topoplot_connect.m), I remove the abs command [plotchans = abs (plotchans);]but program still plot absolute values.
Please suggest how to plot both positive and negative values. If some other program is available to plot connectivity among all channels, please let me know. Any suggestion would be highly appreciable.
Hi, I just tested the script and it looks like the negative values are plotting fine. If you plotted the color bar, it probably always stays between 0 and 1. This is ok, because if you give 3 connection strengths [-1 0 1], then they will be re-mapped to [0 0.5 and 1]. The relative difference between the connection strengths is maintained. Hope this helps.
Hi,
This is fine that relative difference is maintained but I am interested to look both positive and negative values (say red and yellow for positive, blue dark and light for negative).
Hello
Thank you for your function.
Even if in the function there is a comment line
%%% remove infinite and NaN values %%%
I couldn’t run the function with NaNs in the connectStrength matrix. I think that could be useful to get rid of “outliers” such as connectivity values lower than a defined percentage respect to the max connectivity strength. Instead of plotting them as zeros, as it is if you set the strength limits, it would be clearer to delete those values completely for plotting (expecially if using high density EEG) by replacing them with NaNs. I tried to edit the function for this purpose so that it can get rid of NaNs before plotting, and it seems to work. Could I upload it on MathWorks (crediting you of course)?
Many thanks
I can’t do that for my data. solve it for my data set.
Thank you very much!
It is really helpful!